< back to Biology 426 syllabus >
 |
< Biology 426L - Molecular Biology > Macromolecular Projects |
 |
![]()
| To complete this course, you will conduct two, interelated projects that focus on macromolecular structure-function relationships. Both projects involve using Jmol molecular visualization software to illustrate these relationships for a particular molecule involved in a central dogma or signal transduction process. The molecule that you will work on will be determined by you and your professor (DM). Some candidate molecules will be suggested in class. In the first project, you will use Jmol scripting to produce a molecular, web-based tutorial suitable for publishing as an exhibit in CLU's Online Macromolecular Museum (OMM). The second project will involve choosing a particular rendering of your molecule from the first project, saving that rendering as a *.wrl file that can be read as a 3-dimensional model by our rapid protyping printer, and printing a physical model of your molecule in 3-D. You will give oral presentations to the class on your molecule at the end of the semester, using both your web project and your physical model. |
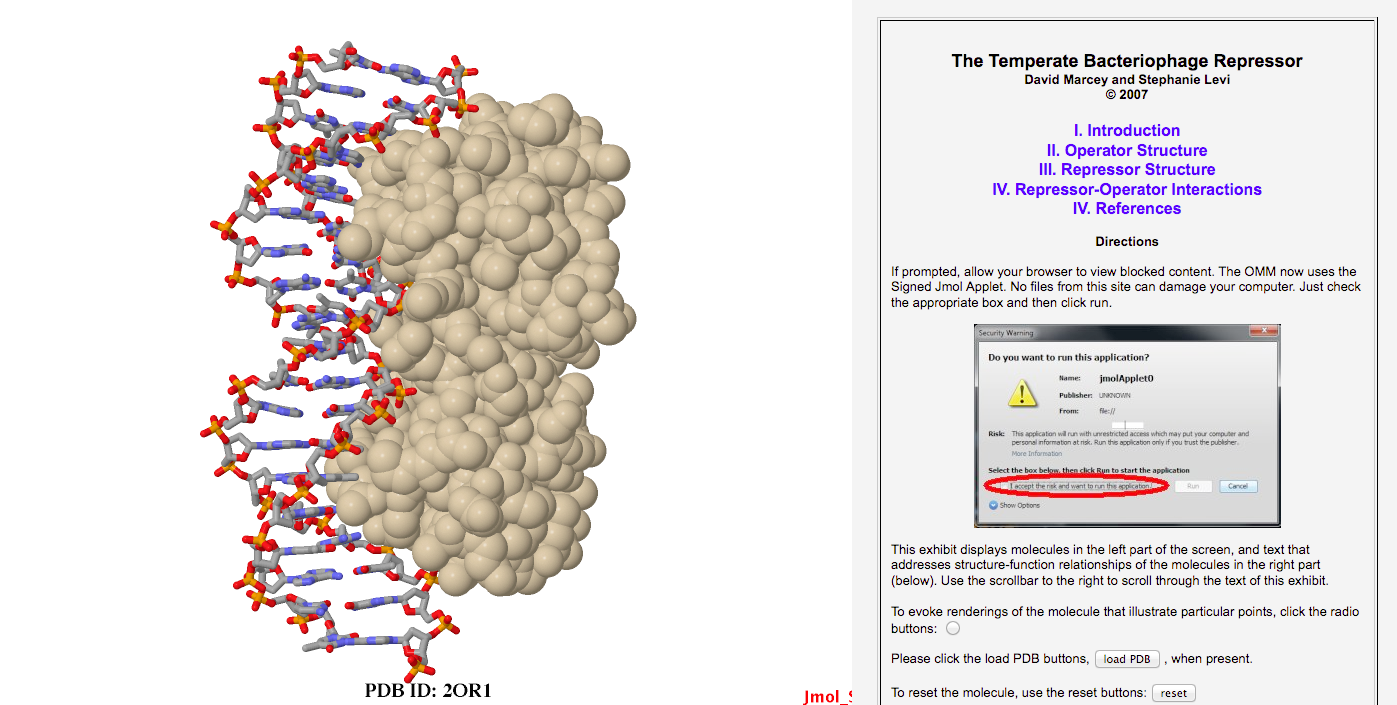
| There are several skills that you will need to master in order to produce the requisite projects. First, you will use Dreamweaver software to build a web page that will display your molecule along with text that describes various features of that molecule. See the exhibits section of the OMM in order to gain an idea of what sucessful projects look like. Note that in a particular exhibit, say the bacteriophage repressor, the buttons in the text frame evoke illustrative renderings of the molecule to the left. You will be given a template web page (*.html file) in order to begin work on your molecule and you will be instructed in how to insert your molecule of choice into the web page and how to use Jmol scripting commands contained in buttons to render the molecule in informative ways. In order to begin learning Jmol scripting commands that you will likely need, you should spend MULTIPLE HOURS going through and studying the Introduction to Jmol Scripting exhibit at the OMM. DM will get you started using this exhibit, using the Jmol applet, and using more advanced Jmol documentation. |
![]()
Jmol Project Links
An Introduction to Jmol Scripting tutorial at CLU's OMM
Companion Website for Herraez's How to Use Jmol...
Jmol training guide at CBM-MSOE
Jmol Quick Reference Sheet (CBM-MSOE)
Jmol Training Guide 1 (CBM-MSOE)
Jmol Training Guide 2 (CBM-MSOE)
Jmol Training Guide 3 (CBM-MSOE)
Physical
Model Design Parameters (CBM-MSOE)
![]()
< back to Biology 426 syllabus >