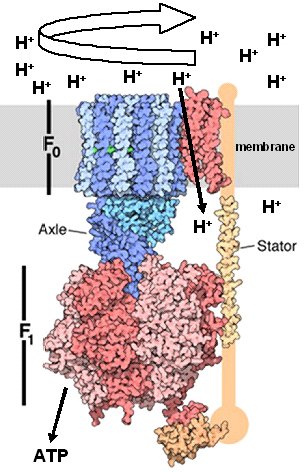
I. IntroductionThe dephosphorylation of adenosine triphosphate (ATP) provides energy for many biochemical reactions. ATP is primarily produced by the enzyme ATP Synthase (ADP + Pi ---> ATP). F-ATP Synthases are found in the inner mitochondrial membranes and the chloroplast thylakoid membranes of eukaryotes, as well as in prokaryotic plasma membranes. ATP synthases are an ancient family of proteins that are highly conserved throughout all kingdoms of life. ATP Synthase functions similarly to the turbines of hydroelectric plants that utilize the kinetic energy of water that flows through dams. In this analogy, a proton gradient is likened to dammed water, and the flow of protons down the gradient is like water driving the turbines of a hydroelectric engine. Like hydroelectric turbines, ATP synthase components rotate in response to the proton flow, and this rotational energy is then coupled to ATP synthesis. These amazing enzymes thus function as molecular engines that harness the energy derived from proton flow to drive phosphorylation of ADP, producing ATP that can be utilized by any number of enzymes to facilitate catalysis of particular biochemical reactions. The F-ATP Synthase includes the Fo rotary motor complex embedded in the membrane, the F1 catalytic complex that synthesizes ATP, and a Stator that connects them and which prevents rotation of the catalytic subunits. The central stalk (axle) is considered part of F1, and its rotation is coupled to that of the membrane rotor (see Figure 1). The Fo rotor spins in response to proton (H+) flow down a concentration gradient across the membrane. This rotation causes the central stalk (axle) to rotate, altering the conformation of components of the F1 base, driving the synthesis of ATP. The synthase can also act as an H+ pump ATPase when its rotations are reversed by ATP hydrolysis.
Shown at left is a partial structure of yeast mitochondrial ATP Synthase with orientation, arrows, and labels corresponding to those of Figure 1 (Dautant, et al., 2010, unpublished -see PDB entry 2WPD). The elegant structure of this molecule allows it to perform its remarkable functions. To understand the functional details linking proton flow, rotation of Fo, and synthesis of ATP by F1, it will be useful to consider the Fo and F1 complexes individually, using the structure of bacterial (E. coli) complexes of Fo as determined by Rastogi and Girvin (1999) and the structure of bovine F1 complexes as determined by Gibbons et al. (2000). II. Fo Structure and Function Links Proton Flow to the Fo RotorIn bacteria, the Fo complex contains the subunits a, b and c, in a ratio of 1a:2b:c10-15. The number of c subunits are fixed within a species, but are variable among different species. Note: Unless otherwise stated, in the following representations the Fo complex will be either be oriented as in Figure 1, above (sideview), or in a top down view (above the membrane), looking through the Fo channel towards the central stalk and F1 complex. In E. coli, Fo consists of an a subunit, a b Stator unit (not shown), and a ring of 12 identical c subunits. The c ring of Fo rotates, while the other components of the complex do not. Each c subunit is a helix-loop-helix, comprising a C-terminal alpha helix and an N-terminal helix. These two helices each span the membrane. Each C-terminal helix contains the important acidic amino acid, Aspartate 61. This residue's sidechain, capable of protonation and deprotonation, plays a major role in the rotation of the c ring. As can be seen, only two of the c ring's C-terminal alpha helices are in close proximity to the a subunit at any one time. These features are important and will be discussed below. The a subunit contains 4 helices. The helix closest to the c ring contains the basic amino acid, Arginine 210. The sidechain of Arg 210 and other residues provide a hydorophillic environment that promotes the deprotenation of Asp 61 as its helix rotates to contact the a subunit. Note: in this top down view, rotation of the c ring is in the clockwise direction. Deprotenation of Asp 61 causes a profound change in the conformation of the C-terminal helix of the deprotonated subunit: it twists along its axis ~140o relative to the N-terminal helix. The twisting of the C terminal helix in response to deprotenation of Asp 61 can be visualized in a morphed simulation using single NMR structures of protonated and deprotonated c subunits (Rastogi and Girvin,1999) as starting and ending points.This PDB file for this simulation was generated using the Yale Morph Server at the Database of Macromolecular Movements, maintained by the Gerstein lab. The twisting of the C terminal helix just described suggests a model in which local rotation of a single c subunit is a major physical force that spins the entire ring, as in a "wheels within wheels" type of mechanism (Rastogi and Girvin,1999). This rotational process of Fo can be visualized in an animation provided by the Girven lab. It is now useful to consider the protonation state of the 12 c subunits in the c ring of Fo. As can be seen, only the Asp 61 of the C-terminal helix that has just rotated into the vicinity of the a subunit's Arginine 210 is in a deprotonated state. The remaining 11 Asp 61s in the other C-terminal helices are protonated. Careful inspection shows that only theC-terminal helix containing the deprotonated Asp 61 is in the twisted (deprotonated) conformation. This has an important consequence for the directional rotation of the c ring. Since the deprotonated Asp 61 is negatively charged, counterclockwise rotation of the c ring is thermodynamically unfavorable, since this would place the deprotonated Asp 61 in the hydrophobic environment of the membrane. Clockwise rotation of the ring is favored, however, as the Asp 61 would still be in the hydrophillic vicinity of Arg 210 and other residues of the a subunit.This position allows it to be reprotonated and enter the membrane environment as the c ring spins in the clockwise direction. Note that as the Fo c ring rotates, the Fo a subunit remains stationary. A side view through a translucently rendered a subunit shows its association with the C-terminal helix containing the deprotonated Asp 61 and the C-terminal helix containing the newly reprotonated Asp 61. The the remaining C-terminal helices are exposed to the hydrophobic membrane. The Arg 210 of the a subunit is seen to reside btween the deprotonated and reprotonated Aspartates. The structure of Fo facilitates the flow of H+ ions down the proton concentration gradient by providing two half-channels for this flow, an entry and an exit channel. These can be visualized in a side view of the c ring through the translucently rendered a subunit:
The elegant mechanism for converting the electrical energy of the proton gradient into rotary (kinetic) energy of c ring spinning may be summarized by following the journey of a proton as it flows across the membrane through Fo:
The E. coli Fo c ring just described has 12 c subunits. Thus, each proton that moves through Fo rotates the ring in 30o steps relative to the stationary components of ATP synthase (12 c's x 30o = 360o). In the next section we will consider how the rotary motion of the Fo c ring powers ATP synthesis in the F1 complex. III. F1 Structure and FunctionShown at left is the F1 complex of the F-ATP synthase from bovine heart mitochondria, oriented with the top pointing toward the membrane bound Fo complex described above (not shown). Also shown is the central stalk (axle), which is linked to the Fo c ring above, and which therefore rotates with that ring. The catalytic F1 complex lies below the central stalk. Unlike the stalk, the catalytic complex is fixed and is prevented from rotating by its binding to the Stator mentioned previously (not shown), which connects to the stationary Fo a subunit (see Figure 1). The central stalk contains the gamma, delta, and epsilon subunits. The catalytic complex is a hexamer alternately packed with three alpha subunits and three beta subunits. Although all subunits of the catalytic complex bind to nucleotides, only the beta subunits are capable of catalyzing the phosphorylation of ADP to produce ATP. The gamma subunit of the stalk is observed to penetrate deep within the catalytic complex, where it engages the beta subunits. Keeping in mind the structural features of F1 just presented, it is now possible to understand the Binding-Change Model that explains how the rotational energy of the central stalk is transduced into the production of ATP. In this model, the gamma subunit of the central stalk sequentially engages the beta subunits of F1 as it rotates in sync with the Fo rotor. This interaction induces conformational changes that drive the release of ATP from these catalytic subunits. Each stationary beta subunit transitions between three conformations. The three conformers of the beta subunits are LOOSE, TIGHT, and OPEN and each beta subunit cycles sequentially between them (L --> T--> O), the cycles being orchestrated by the rotating gamma subunit of the central stalk. The LOOSE conformation permits the loose binding of ADP and Pi substrates, but ATP catalysis does not occur until the beta subunit transitions to the TIGHT conformation. The TIGHT conformation produces ATP (ADP + Pi ---> ATP) but is incapable of releasing this catalytic product. Only when the TIGHT to OPEN conformational change is induced can the beta subunit release ATP. The perspective here is from the bottom of the ATP Synthase, looking up toward the membrane. In this bottom-up view, the rotation of the central stalk gamma subunit is counterclockwise. With each rotation of 120o, the gamma subunit engages a stationary beta catalytic subunit, changing its conformational state as described. The result is the production and release of ATP by F1. Remembering that each 360o rotation of the Fo rotor is linked to a 360o rotation of the central stalk, a simple calculation reveals the stoichiometry of H+ translocation through Fo and ATP production by F1. If there are 12 c subunits in the c ring of Fo (each carrying a proton), and each 360o rotation of the gamma subunit of the central stalk produces 3 ATPs (one for each F1 beta subunit), then the transport of 4 protons are required to generate every ATP (12 protons/rotation of the c ring rotor / 3 ATPs/rotation of the central stalk). Of course, this ratio depends upon the number of c subunits in the Fo rotor, which can vary depending on the ATP Synthase under consideration. Clearly, understanding the structure-function relationships of ATP Synthase gives one a deep appreciation of the power of natural selection in fashioning amazing biomolecular machines!
IV. ReferencesGibbons, C.; et. All. The Structure of the Central Stock in Bovine F1-ATPase at 2.4 Å resolution. Nature Structural Biology. 2000 Nov;7(11):1002-4. Rastogi, V.K.; Girvin, M. Structural Changes Linked to Proton translocation by Subunit C of the ATP Synthase. Nature. 1999 Nov 18;402(6759):247, 249. Stock D.,
Leslie, A.G and Walker, J.E.. Molecular architecture of the
rotary motor in ATP synthase. Science. 1999 Nov 26;286(5445):1700-5. |
||